Archived Content
The National Institute of Mental Health archives materials that are over 4 years old and no longer being updated. The content on this page is provided for historical reference purposes only and may not reflect current knowledge or information.
Genes That Turn On Together Hold Secrets of Brain’s Molecular Instructions
“Guilt by Association” Could Identify Genetic Culprits in Human Illness
• Science Update
For the first time, scientists have mapped groups of genes that turn on together in the human brain, revealing a kind of Rosetta Stone of its molecular organization. These never-before-seen patterns of co-expressed genes hold promise for implicating genetic mechanisms conferring risk for illness through “guilt by association,” say the researchers.
NIMH grantee Daniel Geschwind, M.D., Ph.D., Michael Oldham, and colleagues at the University of California Los Angeles (UCLA), report on their findings in the November 2008 issue of Nature Neuroscience.
Background
Since all cells in the brain start out with the same genes, it’s the particular set of genes that gets expressed – or turned on – that determines the kind of cell it becomes. So a set of genes that turn on together, called a “transcriptome,” holds clues to the molecular instructions underlying the brain’s different cell and tissue types. Using methods developed with collaborator Steve Horvath, Ph.D., the UCLA researchers were able to see a gene’s pattern in relation to those of all other genes. They compared samples of different brain regions (cortex, caudate nucleus and cerebellum) from many deceased individuals, looking for genes that turned on and off together.
Results of This Study
To their surprise, the researchers found many telltale patterns of genes being expressed together that held up across different brain regions, individuals, and experimental methods. These “modules” of co-expressed genes corresponded to brain components with different functions – such as neurons and other types of brain cells.
Significance
Many of the modules reveal the brain’s “cellular building blocks,” say the researchers. Among other things, they can be used to obtain genetic and functional information specific to a particular cell type from whole brain tissue without having to isolate just those cells from intertwining thickets of circuitry. Because genes expressed in the same module likely share similar underlying functions, these modules also provide a shortcut to discovery of the genetic underpinnings of disease through what the researchers call “the principle of guilt by association.” They provide clues to the functions of previously unknown genes that are part of the module. Also, if gene expression patterns from disease-affected individuals differ from the predicted norm, it could mean that those genes or cells are implicated in disease pathways. This could advance understanding of the human brain’s complexity and of the underlying events that contribute to disease processes.
What’s Next?
Efforts are underway to understand how brain disorders might alter the map of the human brain transcriptome. For example, the researchers identified a group of genes that turn on together in a part of the brain where new neurons continue to be born in adults. Understanding the regenerative capacity of such cells could lead to a strategy for treating neurodegenerative conditions such as Alzheimer’s disease.
In-Sync
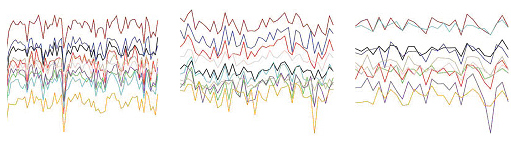
The same genes turn on and off together, but according to different instructions, in samples from three different tissues of the human brain, cortex (left), caudate nucleus (center), cerebellum (right). Each color represents the expression pattern of a single gene. These sets of co-expressed genes, or “modules,” hold secrets to the human brain’s molecular instructions.
